Archives
We then went on to
We then went on to see if these proteins bound non-specifically to an order JWH 073 of affinity resins; FLAG, TALON, Calmodulin and IgG and identified hundreds of non-specific interactors for each resin. We compared these lists to identify which proteins bind to multiple resins non-specifically. We refer to this comparative list as the DT40 beadome (Supplementary Table 2) and again performed GO annotation to characterise these proteins (Supplementary Fig. 2).
After establishing the nature of potential non-specific contaminants in AP experiments, we then used a C-terminal tandem 3x FLAG-2x (HIS6) tagged Phosphatidyl inositol 5-phosphate 4-kinase 2β protein (PI5P4K2β) (JPR3 cell line) and a C-terminal tandem calmodulin binding protein (CBP) and a-Protein-A tagged FANCC allele (FANCC cell line) to investigate their interacting partners. We coupled this with SILAC labelling to make analysis quantitative (qAP-MS), thus enabling us to discriminate genuine from non-specific interactors. It is already known that the nuclear membrane protein PI5P4K2β forms homodimers and also heterodimerises with PI5P4K2α with no other proteins interacting, not surprisingly, as its substrates are lipids. The MS identified interactors and isotopic ratios are presented as Supplementary Table 3A and B for FLAG and TALON affinity purifications respectively.
The Fanconi complex however is less well understood and interactors are transient and of low abundance upon stalling of replication forks. We therefore wished to investigate this complex more thoroughly and identified some new interactors of which a couple were independently confirmed (genetically) by another group. The resulting partners and their quantitative analysis are presented in Supplementary Table 4.
Experimental design, materials and methods
In initial studies to identify abundant DT40 proteins, three replicates of 106 total cells, grown in RPMI media with l-glutamine supplemented with 10% FBS and 1% chicken serum (CS) (all GIBCO) and maintained at 37°C at 5% CO2, as described previously [2], were lysed in 10ml lysis buffer, comprising 1× PBS (pH7.4) 1% Triton X-100, 1mM PMSF and 1.5× EDTA free protease inhibitor cocktail (Roche), on ice and the cleared supernatants separated by reducing SDS-PAGE and stained. All bands were excised and the sample prepared and analysed by MS as described below.
For DT40 beadomes and SILAC-iPAC experiments all cell lines were grown in parallel to 108 total cells. Cells were lysed in 10ml lysis buffer on ice and the cleared supernatants quantified, then added to 100μl prewashed resins. For SILAC-iPAC experiments quantified lysates were mixed 1:1 (typically 5mg) then added to 100μl prewashed resins. JPR3 and wt DT40 cell extracts were purified using EZview ANTI-FLAG M2 Affinity Gel (Sigma) and TALON Metal Affinity Resin (Clontech). FANCC and wild-type DT40 cell extracts were purified with Calmodulin Sepharose 4B and IgG Sepharose 6 (both GE Healthcare). Native protein complexes were allowed to bind for 1h then non-bound proteins were removed by centrifugation at 2000g, the resins were washed 3 times for 15min in 1ml lysis buffer and the bound proteins were eluted in 100μl of elution buffer. Elution buffers were lysis buffer pH 7.4 containing either 100μg/ml 3× FLAG peptide for FLAG resin, 150mM imidazole for TALON resins, or 200mM EDTA for calmodulin resins. 100mM glycine pH2.5 was used for IgG resins.
Acknowledgements
We thanks Prof. R. Irvine for providing the JPR3 cell line, to Dr. E. Rajendra for providing the FANCC cell line, FANC antibodies and helpful discussions, to Dr. M. Deery and J. Howard for assistance with MS. This work was funded by the Biotechnology and Biological Sciences Research Council (UK) Grant BB/H024085/1.
Value of the data
Experimental design, materials and methods
Genome-wide association studies (GWAS) focus on panels of genetically diverse lines and exploit historical recombination between loci. Where recombi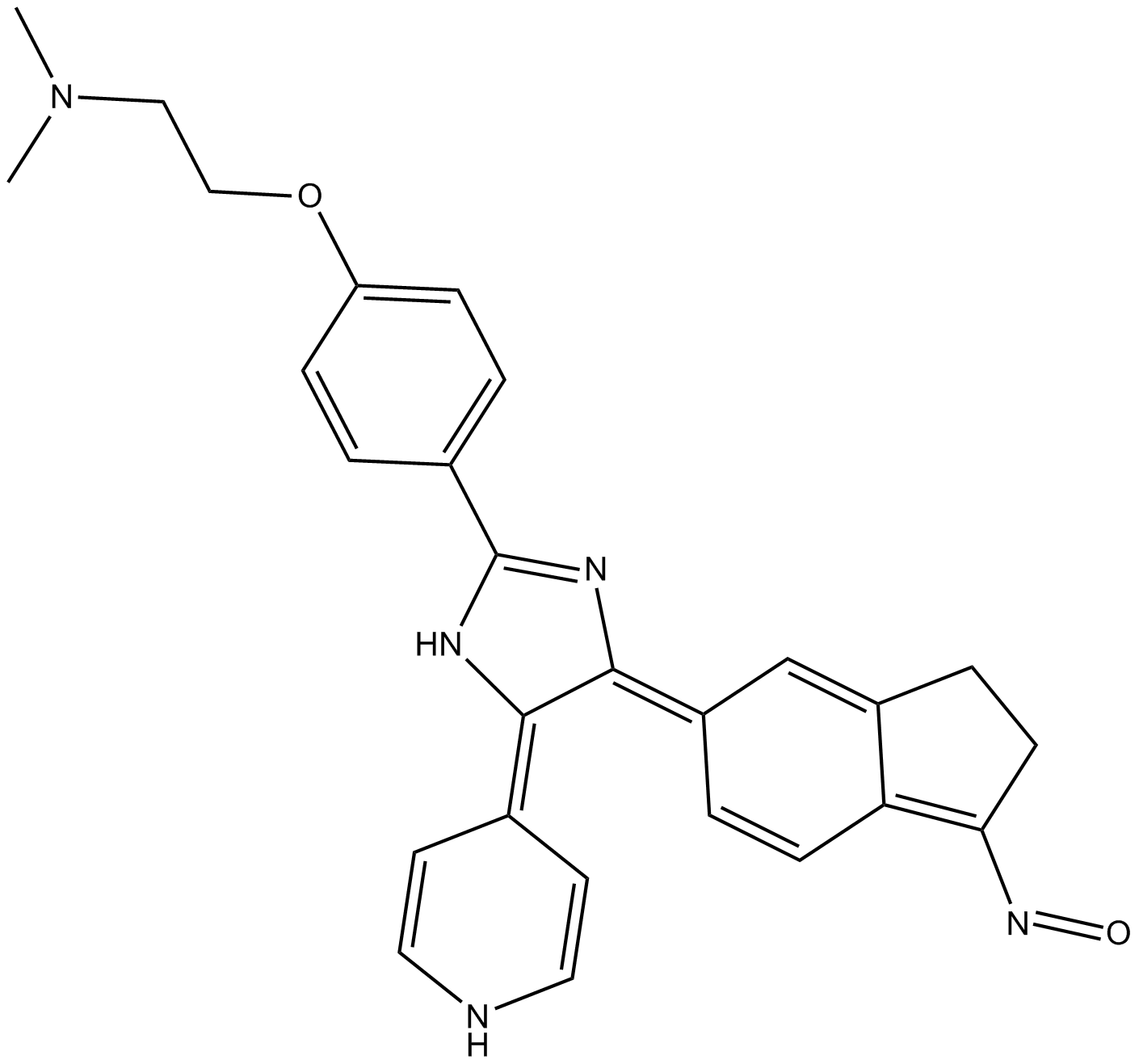 nation between loci is observed rarely, those loci are said to be in Linkage Disequilibrium (LD). The approach of identifying markers in LD with loci controlling traits is an important tool in human genetics studies and has been applied successfully in plants with well-established genome resources: maize, rice and Arabidopsis thaliana[1–5]. In crop science, GWAS can be used to understand the precise molecular basis of trait variation by identifying causative mutations, or at least to provide tightly linked markers to aid marker-assisted selection. To overcome the problem of lack of genome sequence resources, mRNAseq-based approaches have been developed for the polyploid crop species Brassica napus that enable high throughput identification of single nucleotide polymorphism (SNP) markers [6] and high density linkage map construction [7]. The linkage map enabled the development of a genomic framework; comprising genome sequence scaffolds from the progenitor species of B. napus (Brassica rapa and Brassica oleracea) rearranged into “pseudomolecules” representative of genome organisation of B. napus, which were then used successfully to underpin GWAS studies [8].
nation between loci is observed rarely, those loci are said to be in Linkage Disequilibrium (LD). The approach of identifying markers in LD with loci controlling traits is an important tool in human genetics studies and has been applied successfully in plants with well-established genome resources: maize, rice and Arabidopsis thaliana[1–5]. In crop science, GWAS can be used to understand the precise molecular basis of trait variation by identifying causative mutations, or at least to provide tightly linked markers to aid marker-assisted selection. To overcome the problem of lack of genome sequence resources, mRNAseq-based approaches have been developed for the polyploid crop species Brassica napus that enable high throughput identification of single nucleotide polymorphism (SNP) markers [6] and high density linkage map construction [7]. The linkage map enabled the development of a genomic framework; comprising genome sequence scaffolds from the progenitor species of B. napus (Brassica rapa and Brassica oleracea) rearranged into “pseudomolecules” representative of genome organisation of B. napus, which were then used successfully to underpin GWAS studies [8].